Association between Apolipoprotein E Polymorphism and Chronic Kidney Disease in the Korean General Population: Dong-gu Study
Article information
Abstract
Background
Few studies have investigated the association between Apolipoprotein E (APOE) polymorphisms and chronic kidney disease (CKD) in the general population, and their results are inconsistent.
Methods
The current study population was composed of 9,033 subjects aged ≥ 50 years who participated in the baseline survey of the Dong-gu Study, which was conducted in Korea between 2007 and 2010. APOE polymorphisms were identified by polymerase chain reaction, and the estimated glomerular filtration rate (eGFR) was calculated using the Modification of Diet in Renal Disease equation.
Results
Individuals with the APOE E2 allele had significantly lower total and low density lipoprotein cholesterol levels, those with the APOE E4 allele had lower high density lipoprotein (HDL) cholesterol levels, and those with the APOE E3 allele had lower log-triglyceride levels. Adjusting for covariates (sex, age, body mass index, smoking, systolic blood pressure, hypertension, diabetes, total cholesterol, HDL cholesterol, log-transformed triglycerides, and log-transformed albumin to creatinine ratio), mean eGFR was not significantly different among APOE alleles (E2, 69.4 mL/min/1.73 m2; E3, 69.5 mL/min/1.73 m2; E4, 69.4 ml/min/1.73 m2; P = 0.873). Additionally, the odds ratios (ORs) indicated that APOE polymorphisms were not independent risk factors for CKD (OR, 1.07; 95% confidence interval [CI], 0.91 to 1.26 for the E2 vs. E3 allele; OR, 1.01; 95% CI, 0.88 to 1.16 for the E4 vs. E3 allele).
Conclusion
APOE polymorphisms were not associated with either eGFR or CKD in the general Korean population.
INTRODUCTION
Apolipoprotein E (APOE) is a key component in lipoprotein complexes and has a central role in cholesterol and triglyceride metabolism.1,2,3) The APOE gene has three common alleles (E2, E3, and E4) that arise from two single nucleotide polymorphisms in exon 4, producing six possible genotypes (E2E2, E2E3, E2E4, E3E3, E3E4, and E4E4).4) APOE isoforms differ in their receptor binding ability, as the APOE E2 allele has a cholesterol-lowering effect and the APOE E4 allele has a cholesterol-raising effect compared with the APOE E3 allele.5) Thus, APOE polymorphisms are expected to play a major role in the pathogenesis and progression of a variety of renal diseases.6) However, there are discrepancies in the literature regarding the role of APOE polymorphisms in the genetic predisposition to kidney disease. Two studies provide evidence that the APOE E2 allele confers a genetic risk,7,8) whereas others indicate that the APOE E4 allele is a risk factor for the progression of renal failure.9,10) Yet others demonstrate no association between APOE polymorphisms and kidney disease.11,12)
The aim of the present study was to evaluate the association between APOE polymorphisms, estimated glomerular filtration rate (eGFR), and chronic kidney disease (CKD) in a Korean general population aged ≥ 50 years.
METHODS
1. Study Population and Data Collection
The Dong-gu Study is an ongoing prospective study to investigate the prevalence, incidence, and risk factors for chronic disease in an urban population. National resident registration records were used to identify potential participants. In total, 34,040 eligible subjects of ≥ 50 years of age who resided in the Dong-gu district of Gwangju Metropolitan City in South Korea were invited to participate by mail and telephone from 2007 to 2010. In total, 9,260 subjects (27.2% response rate; 3,711 men and 5,549 women) were enrolled. For the present study, 109 subjects were excluded due to missing data (for genotype, blood lipids, serum creatinine, urine albumin, or life-style), 118 subjects with the E2E4 genotype were excluded, and data from the remaining 9,033 participants (3,610 men and 5,423 women) were included. This study was conducted in accordance with the Declaration of Helsinki guidelines, and the study protocol was approved by the institutional review board of Chonnam National University Hospital. Informed consent was obtained from each subject.
Information about medical history and lifestyle characteristics was obtained from all subjects using standardized questionnaires. Smoking status was classified as non-smoker, former smoker, or current smoker. Height and waist circumference were measured to the nearest 0.1 cm. Weight was measured to the nearest 0.1 kg, and body mass index (BMI) was calculated by dividing weight (kg) by height squared (m2). Diabetes was diagnosed by either a fasting plasma glucose ≥ 126 mg/dL or the use of insulin or other hypoglycemic medications. Blood pressure was measured on the right upper arm using a mercury sphygmomanometer (Baumanometer; WA Baum Co., Inc., Copiague, NY, USA) with the appropriately sized cuff after subjects were seated for at least 5 minutes. Three consecutive measurements of systolic blood pressure (SBP) and diastolic blood pressure (DBP) were performed at 1-minute intervals, and the average was used for analysis. Hypertension was defined as a SBP ≥ 140 mm Hg or a DBP ≥ 90 mm Hg, or the use of blood pressure-lowering medications.
Blood samples were drawn from an antecubital vein after a 12-hour overnight fast. Serum was separated within 30 minutes and stored at -70℃ until analysis. Serum total cholesterol, high density lipoprotein (HDL) cholesterol, triglycerides, and fasting blood glucose levels were measured by enzymatic methods using an automated chemical analyzer (model 7600; Hitachi Ltd., Tokyo, Japan).
2. Apolipoprotein E Genotyping
Genomic DNA was extracted from peripheral blood using either an AccuPrep Genomic DNA Extraction Kit (Bioneer, Seoul, Korea) or a QIAamp DNA Mini Kit (Qiagen Inc., Chatsworth, CA, USA) according to the manufacturer's protocol. APOE genotypes were determined using the polymerase chain reaction as described by Hixson and Vernier,13) with a slight modification. The APOE genotyping method used in this study has been reported previously.14)
3. Estimation of Glomerular Filtration Rate and Chronic Kidney Disease
Kidney function was assessed by calculating the eGFR using the Modification of Diet in Renal Disease equation as follows:15)
CKD was defined as eGFR of <60 mL/min/1.73 m2 according to the National Kidney Foundation Kidney Disease Outcomes Quality Initiative guidelines.16)
4. Statistical Analysis
Data are presented as either means ± SD or percentages for categorical variables. Analysis of variance and Pearson's chi-square tests were used to compare baseline characteristics across APOE genotypes. A post-hoc analysis with Tukey's test was also completed for all possible paired comparisons. Triglyceride and albumin to creatinine ratio (ACR) levels were not normally distributed. Thus, they were log-transformed prior to analysis to approximate a normal distribution, and geometric means with 95% confidence intervals (CI) are presented. The six possible APOE genotypes (E2E2, E2E3, E2E4, E3E3, E3E4, and E4E4) were categorized into three groups for analytical purposes: APOE E2 (E2E2 and E2E3), APOE E3 (E3E3), and APOE E4 (E3E4 and E4E4). Subjects with the E2E4 genotype were excluded due to the potentially opposing biological effects of the E2 and E4 alleles. Analysis of covariance was used to compare mean eGFR values according to APOE allele. Model I was adjusted for age and sex. Model II included the Model I variables plus BMI, smoking, SBP, hypertension, diabetes, total cholesterol, HDL cholesterol, log-transformed triglycerides, and log-transformed ACR. Logistic regression was used to calculate odds ratios (ORs) for CKD according to APOE allele. Statistical analyses were performed using the IBM SPSS ver. 20.0 (IBM Co., Armonk, NY, USA), and P-values of <0.05 were considered to indicate a significant difference.
RESULTS
1. General Characteristics of the Subjects according to Apolipoprotein E Allele
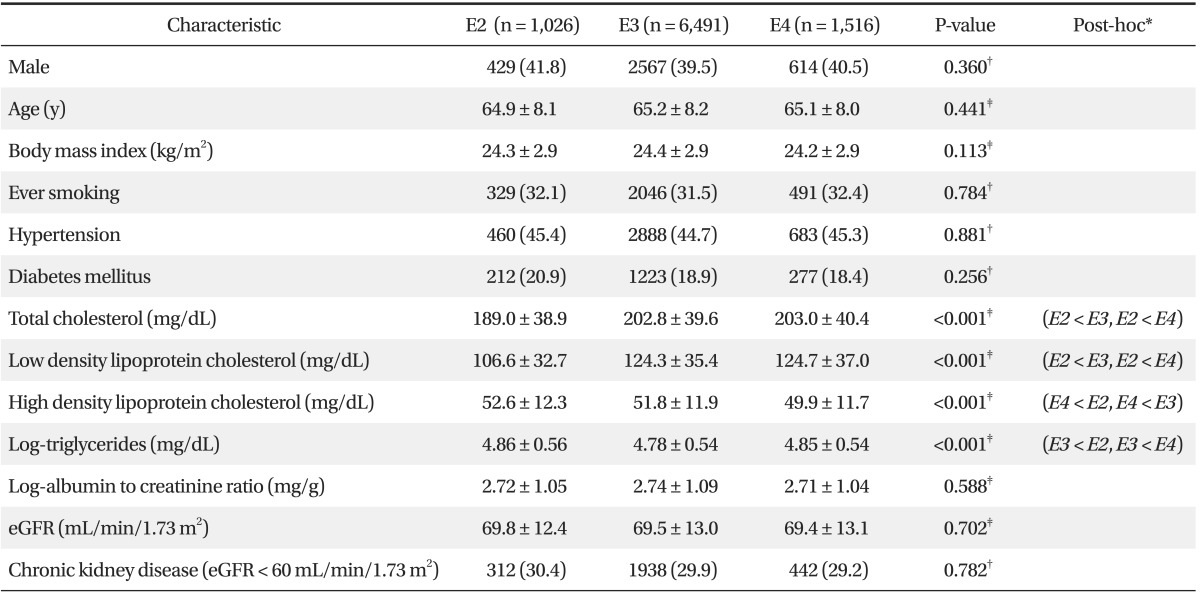
Table 1 shows the general characteristics of the subjects and their relationships with APOE alleles. Participants with the APOE E2 allele had significantly lower total and low density lipoprotein (LDL) cholesterol levels than those with the APOE E3 or E4 allele (P < 0.001). Participants with the APOE E3 allele had significantly lower log-triglyceride levels (P < 0.001) than those with the APOE E2 or E4 allele. Participants with the APOE E4 allele had significantly lower HDL cholesterol levels (P < 0.001) than those with the APOE E2 or E3 allele.
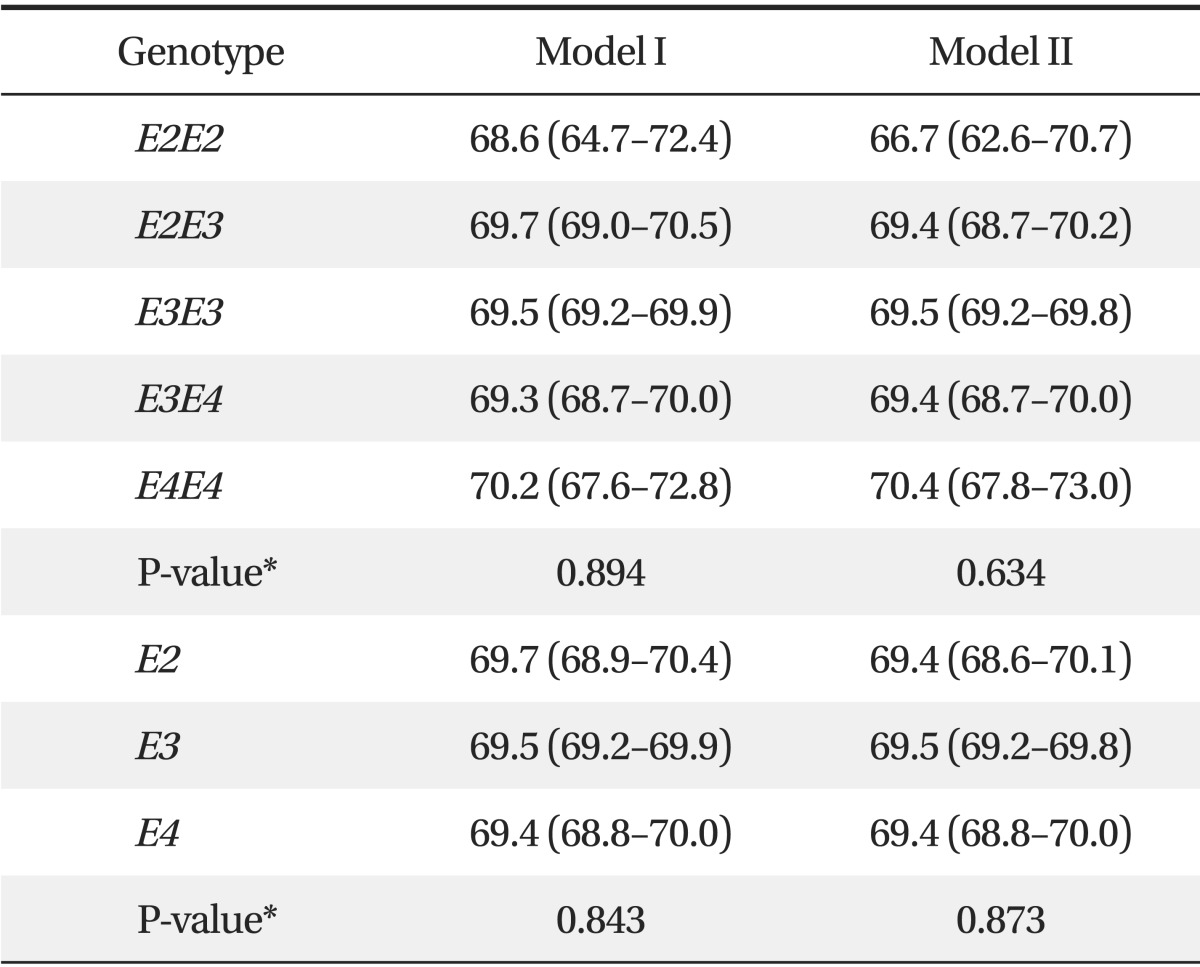
2. Comparison of Mean Estimated Glomerular Filtration Rate according to Apolipoprotein E Allele
Table 2 shows the mean eGFR values (± SD) grouped according to APOE allele. When adjusted for additional covariates (sex, age, BMI, smoking status, SBP, hypertension, diabetes, total cholesterol, HDL cholesterol, log-transformed triglycerides, and log-transformed ACR), mean eGFR was not significantly different among the APOE alleles (APOE E2, 69.7 mL/min/1.73 m2; 95% confidence interval, 68.9 to 70.4 mL/min/1.73 m2; APOE E3, 69.5 mL/min/1.73 m2; 95% confidence interval, 69.2 to 69.9 mL/min/1.73 m2; APOE E4, 69.4 mL/min/1.73 m2; 95% confidence interval, 68.8 to 70.0 mL/min/1.73 m2; P = 0.873).
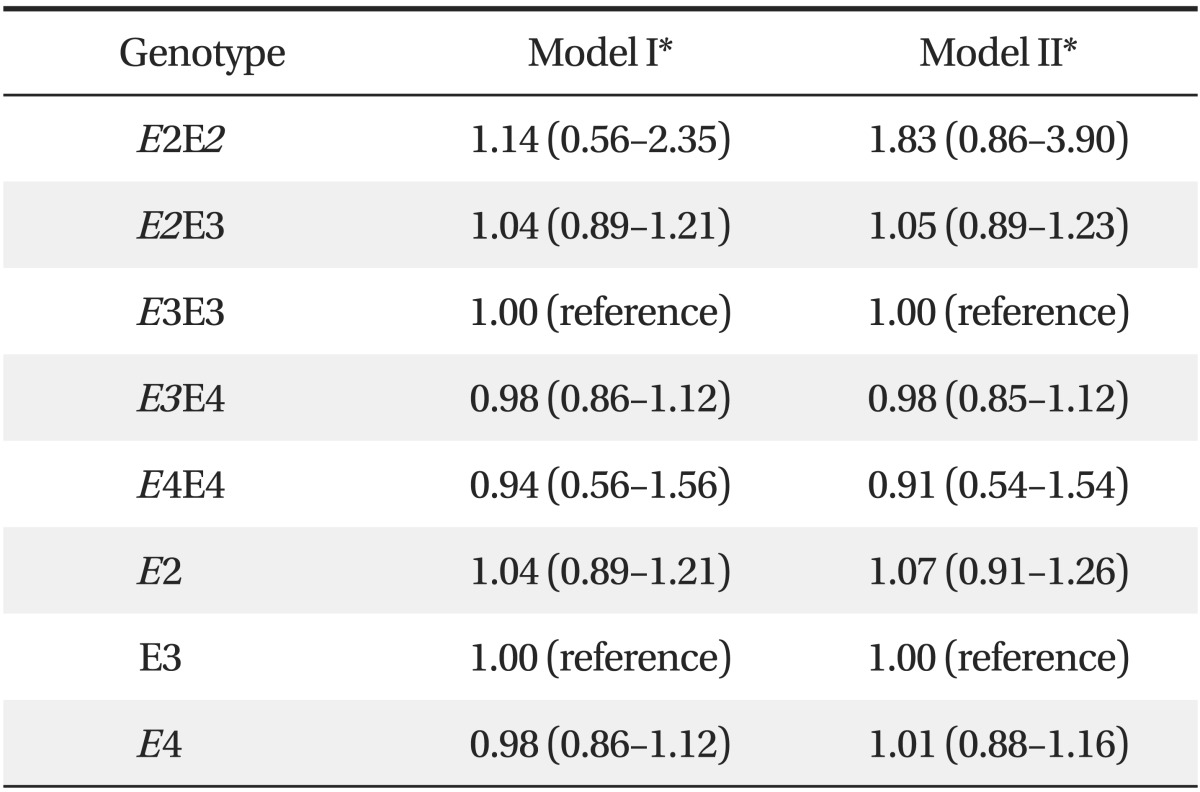
3. Odds Ratios for Chronic Kidney Disease according to Apolipoprotein E Allele
Table 3 lists the ORs for CKD according to APOE allele. When adjusted for albuminuria and other covariates (sex, age, BMI, smoking, SBP, hypertension, diabetes, total cholesterol, HDL cholesterol, log-transformed triglycerides, and log-transformed ACR), the ORs indicated that the APOE polymorphism was not an independent risk factor for CKD (OR, 1.07; 95% CI, 0.91 to 1.26 for the APOE E2 vs. E3 allele; OR, 1.01; 95% CI, 0.88 to 1.16 for the APOE E4 vs. E3 allele).
DISCUSSION
We evaluated the relationship between APOE polymorphisms and CKD in Korean subjects aged ≥ 50 years. Our results demonstrate that individuals with the APOE E2 allele had significantly lower total and LDL cholesterol levels, those with the APOE E4 allele had significantly lower HDL cholesterol levels, and those with the APOE E3 allele had significantly lower log-triglyceride levels. Additionally, eGFR estimates were not significantly different among subjects with different APOE alleles, and the APOE polymorphisms were not independent risk factors for CKD in this population.
Participants with the APOE E2 allele showed significantly lower total and LDL cholesterol levels than those with the APOE E3 or E4 allele. This finding is consistent with previous studies.14,17,18) The inability of APOE E2 isoforms to effectively bind LDL and APOE receptors decreases the amount of very LDL (VLDL), VLDL remnants, and cholesterol from chylomicrons entering hepatocytes. This results in upregulation of the LDL receptor and a subsequent decrease in serum Apolipoprotein B-containing lipoproteins.5) Moreover, LDL clearance likely increases because LDL particles have a greater affinity for the LDL receptor than remnant lipoproteins carrying the defective APOE E2 allele.5)
In contrast to the lack of a correlation between APOE alleles and LDL cholesterol levels, the associations between APOE alleles and HDL cholesterol and triglyceride levels are not entirely clear. In the present study, participants with the APOE E4 allele had the lowest HDL cholesterol levels in accordance with previous reports.14,17) However, other researchers have reported no association between APOE alleles and HDL cholesterol levels.19,20) Many investigators have demonstrated that participants with the APOE E3 allele have the lowest triglyceride levels,14,18,20) similar to our results. However, another study reported that female subjects with the APOE E2 allele have the lowest triglyceride levels.21) Gene-environment interactions may have contributed to the discrepancies between studies. HDL cholesterol levels are modulated by smoking, alcohol consumption, physical activity, and diet.22,23) In another Korean study,14) the associations between the APOE alleles and HDL cholesterol levels were affected by age, and the associations between APOE alleles and triglyceride levels were affected by smoking status.
In the present study, eGFR did not differ significantly among the APOE alleles, and APOE polymorphisms were not independent risk factors for CKD. Several studies have investigated the relationship between APOE alleles and renal function. However, most of these studies were conducted on patients with diabetes7,8,9,10,11,12) and sample sizes were small. Moreover, studies in general populations have not demonstrated consistent results. Some studies have reported a significant association between the APOE E2 allele and CKD,19,20) whereas others indicate that the APOE E4 allele is associated with a decreased risk of CKD.24,25) Yet others have reported no association between APOE polymorphisms and the prevalence of CKD.26) The reason for these conflicting results is poorly understood, but we offer potential explanations. First, the association between the APOE E2 or E4 alleles and CKD in previous studies was significant only in Caucasian participants. In the Atherosclerosis Risk in Communities (ARIC) study,24) the association between the APOE E4 allele and CKD was significant in Caucasian Americans but not in African Americans. In the National Health and Nutrition Examination Survey data,26) a significant association between the APOE E4 allele and CKD was not observed in Mexican-Americans. Second, the study protocols were different in each of these previous studies, and each study used a different method of categorizing APOE groups. In the Cardiovascular Health Study,25) APOE E2E4 was grouped with the APOE E3 alleles. In the present study, APOE E2E4 was excluded due to the potential opposing biological effects of the APOE E2 and APOE E4 alleles. Only APOE E2E3 and APOE E3E3 alleles were compared in the RENASTUR study with a Spanish Caucasian population.20) Moreover, the definition of CKD24,25) and the methods for estimating GFR19,26) were different in each study.
We do not know why APOE polymorphism affected lipid profiles but not eGFR or CKD in our Korean study participants. One potential explanation is that gene-environment interactions may contribute to this discrepancy.14) Another explanation is that a low allele frequency within the study population resulted in an underpowered analysis that did not detect significant associations. In the ARIC study,24) the APOE E2 allele frequency was 14.2% and that of the APOE E4 allele was 28.6%. In the Cardiovascular Health Study,25) the frequencies of the APOE E2 and E4 alleles were 13.7% and 23.0%, respectively. In the present study, the APOE E2 allele frequency was 11.4% and the APOE E4 allele frequency 16.8%. Finally, APOE polymorphisms may not be associated with or minimally affect the kidney function of Korean study participants.
The present study had some limitations. First, the study was cross-sectional. Second, no direct eGFR measurements were obtained. The gold standard for measuring GFR is inulin clearance, but this was not feasible in a large epidemiologic study. Third, medication history, including that of anti-cholesterol medication, was not obtained for all participants.
In conclusion, individuals with the APOE E2 allele showed significantly lower total and LDL cholesterol levels, those with the APOE E4 allele had significantly lower HDL cholesterol levels, and those with the APOE E3 allele had significantly lower log-triglyceride levels. Additionally, eGFR was not significantly different among APOE alleles, and APOE polymorphisms were not independent risk factors for CKD in this population.
ACKNOWLEDGMENTS
This study was supported by a research fund from Chosun University, 2013.
Notes
No potential conflict of interest relevant to this article was reported.